MRM Insights: MYSM1 in Epigenetic Regulation of Mammalian Hematopoiesis

Jooeun (June) Kim

HanChen Wang

Dr. Anastasia Nijnik
Every month, in MRM Insights, a member of the MRM Network is writing about stem cells and regenerative medicine from a different perspective. This month, Dr. Anastasia Nijnik, Associate Professor in the Department of Physiology at McGill University and her lab members HanChen Wang (Research Assistant) and June Kim (Master candidate) are discussing the role of MYSM1 in epigenetic regulation of mammalian hematopoiesis.
MYSM1 in Epigenetic Regulation of Mammalian Hematopoiesis: from Mouse Models to Human Patients
The blood and immune system have some of the highest rates of cell turnover in the human body, with hundreds of billions of new cells produced daily. The process of blood and immune cell production is known as hematopoiesis and is sustained by the hematopoietic stem and progenitor cells (HSPCs) in our bone marrow. The differentiation of HSPCs into the different blood and immune cell lineages requires precise changes in gene expression and is controlled by many transcription factors, chromatin modifying enzymes, and other epigenetic regulators. Dysregulation in the epigenetic mechanisms controlling blood and immune cell production can cause major human diseases, including leukemias, anemias, immunodeficiencies, and bone marrow failures, however our understanding of such mechanisms remains incomplete.
The International Knockout Mouse Consortium (IKMC) was established shortly after the publication of the first human and mouse genome assemblies, with the aim of characterizing the functions of all mammalian protein-coding genes1, 2. Continuing its work as the International Mouse Phenotyping Consortium (IMPC), it is a vital resource for biomedical science, carrying major repositories of knockout mouse strains, mouse embryonic stem cells (ESCs), and phenotypic data that describes the effects of loss-of-function of over 7,500 genes on mammalian physiology.
I first encountered the protein Myb-like SWIRM and MPN domains 1 (MYSM1)3 as a visiting postdoctoral fellow working with IKMC/IMPC at the Wellcome Trust Sanger Institute in Cambridge (UK), now over 10 years ago. At that time, MYSM1 was known as a nuclear protein with deubiquitinase catalytic activity that removes the repressive epigenetic mark histone H2A-K119ub from chromatin4, 5. However, the roles of MYSM1 in vivo in the regulation of mammalian development and physiology were unknown.
Over the past 10 years at the Department of Physiology of McGill University, my research team has characterized MYSM1 as a major regulator of HSPC function and mammalian hematopoiesis3, drawing on the resources of IKMC/IMPC throughout our work. Our contributions include characterizing the functions of MYSM1 across different blood and immune cell lineages using conditional knockout mouse models, and demonstrating that the loss of blood cells in MYSM1-deficiency is mediated by p53 and its downstream stress response pathways3, 6-10. Importantly, in recent years, mutations in MYSM1 were also observed in rare human patients with anemia, immunodeficiency, and bone marrow failure3, indicating the conservation of MYSM1 function between human and mouse and encouraging us to expand our work with clinical collaborations.
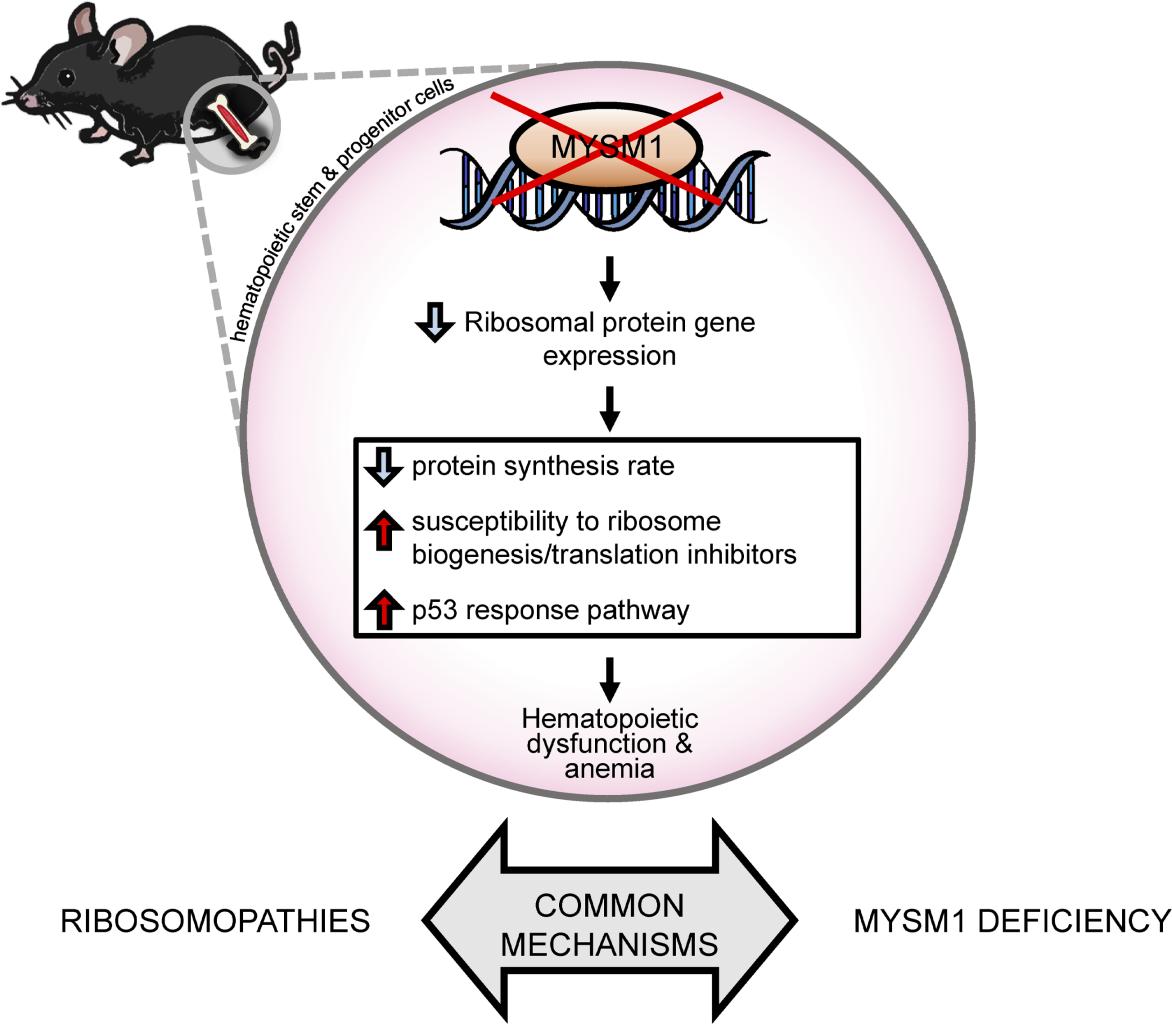
Figure 1. Mechanisms of Hematopoietic Dysfunction in MYSM1 Deficiency; (originally published in JCI Insight. 2020;5(13):e125690).
As MYSM1 is an epigenetic regulator, characterizing the genes directly regulated by MYSM1 in mammalian HSPCs has been a major long-term goal for my research team. This work was conducted in collaboration with Dr. David Langlais (McGill University, Genome Centre), and provided unexpected insights into the molecular mechanisms linking the loss of MYSM1 to bone marrow failure11. The study revealed that MYSM1 maintains the expression of genes encoding the proteins of the ribosome in mouse HSPCs, with loss of MYSM1 resulting in downregulation of these genes, reduction in cellular protein synthesis rates, and the activation of p53, likely via the ribosomal biogenesis checkpoint mechanisms11, 12. This led us to the unexpected conclusion that MYSM1 deficiency may be classified as a form of ribosomopathy, with similar molecular mechanisms leading to hematopoietic dysfunction as in Diamond-Blackfan Anemia (DBA)13. Indeed in recent studies, some patients diagnosed with DBA had been shown to carry mutations in MYSM114, further supporting our conclusions.
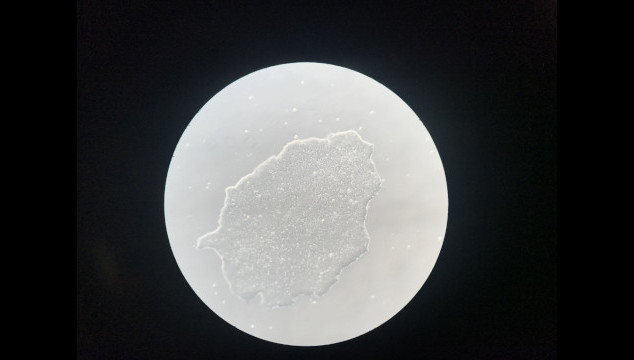
- Figure 2.Growing human iPSCs for the study of MYSM1 functions in human hematopoiesis; (image by HanChen Wang, McGill University).
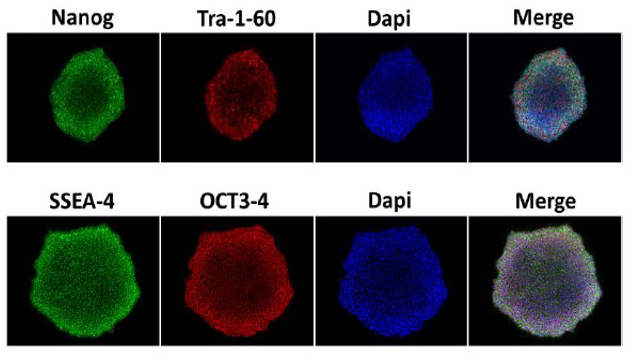
- Figure 3.Staining of MYSM1-deficient human iPSCs for pluripotency markers; (image courtesy of Carol Chen and Dr. Tom Durcan, Early Drug Discovery Unit of the Montreal Neurological Institute-Hospital).
An important aim of our ongoing work is to validate the molecular mechanisms linking MYSM1 deficiency to hematopoietic dysfunction in a human system. With the support of the McGill Regenerative Medicine network, and in partnership with the Early Drug Discovery Unit of the Montreal Neurological Institute, and the Charles-Bruneau Plateforme de Reprogrammation de Cellules of the Centre Hospitalier Universitaire Sainte-Justine, we have established a set of human induced pluripotent stem cells (iPSCs), carrying MYSM1 loss of function mutations. Our ongoing work with these iPSCs promises to advance the understanding of the mechanisms of bone marrow failure in MYSM1 deficiency in humans.
Another goal of our ongoing work is to explore how MYSM1 works to maintain ribosomal gene expression, and our findings indicate a cooperation between MYSM1 and the oncogenic transcription factor cMYC. Importantly, we further showed that the loss of MYSM1 inhibits cMYC oncogenic activity and protects against cMYC-driven lymphoma onset and progression in mouse models15. In future work, with the support of The Leukemia and Lymphoma Society of Canada, we hope to develop these studies to explore MYSM1 as a possible drug target for cMYC driven hematologic malignancies.
References
1. Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 2011, 474(7351): 337-342.
2. Bradley A, Anastassiadis K, Ayadi A, Battey JF, Bell C, Birling MC, et al. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome 2012, 23(9-10): 580-586.
3. Fiore A, Liang Y, Lin YH, Tung J, Wang H, Langlais D, et al. Deubiquitinase MYSM1 in the Hematopoietic System and beyond: A Current Review. Int J Mol Sci 2020, 21(8).
4. Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, et al. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell 2007, 27(4): 609-621.
5. Belle JI, Nijnik A. H2A-DUBbing the mammalian epigenome: expanding frontiers for histone H2A deubiquitinating enzymes in cell biology and physiology. The International Journal of Biochemistry & Cell Biology 2014, 50: 161-174.
6. Nijnik A, Clare S, Hale C, Raisen C, McIntyre RE, Yusa K, et al. The critical role of histone H2A-deubiquitinase Mysm1 in hematopoiesis and lymphocyte differentiation. Blood 2012, 119(6): 1370-1379.
7. Belle JI, Langlais D, Petrov JC, Pardo M, Jones RG, Gros P, et al. p53 mediates loss of hematopoietic stem cell function and lymphopenia in Mysm1 deficiency. Blood 2015, 125(15): 2344-2348.
8. Belle JI, Petrov JC, Langlais D, Robert F, Cencic R, Shen S, et al. Repression of p53-target gene Bbc3/PUMA by MYSM1 is essential for the survival of hematopoietic multipotent progenitors and contributes to stem cell maintenance. Cell Death Differ 2016, 23(5): 759-775.
9. Forster M, Farrington K, Petrov JC, Belle JI, Mindt BC, Witalis M, et al. MYSM1-dependent checkpoints in B cell lineage differentiation and B cell-mediated immune response. J Leukoc Biol 2017, 101(3): 643-654.
10. Forster M, Boora RK, Petrov JC, Fodil N, Albanese I, Kim J, et al. A role for the histone H2A deubiquitinase MYSM1 in maintenance of CD8(+) T cells. Immunology 2017, 151(1): 110-121.
11. Belle JI, Wang H, Fiore A, Petrov JC, Lin YH, Feng CH, et al. MYSM1 maintains ribosomal protein gene expression in hematopoietic stem cells to prevent hematopoietic dysfunction. JCI Insight 2020, 5(13).
12. Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell 2009, 16(5): 369-377.
13. Ito E, Konno Y, Toki T, Terui K. Molecular pathogenesis in Diamond-Blackfan anemia. Int J Hematol 2010, 92(3): 413-418.
14. Ulirsch JC, Verboon JM, Kazerounian S, Guo MH, Yuan D, Ludwig LS, et al. The Genetic Landscape of Diamond-Blackfan Anemia. Am J Hum Genet 2019, 104(2): 356.
15. Lin YH, Wang H, Fiore A, Forster M, Tung LT, Belle JI, et al. Loss of MYSM1 inhibits the oncogenic activity of cMYC in B cell lymphoma. J Cell Mol Med 2021, 25(14): 7089-7094.
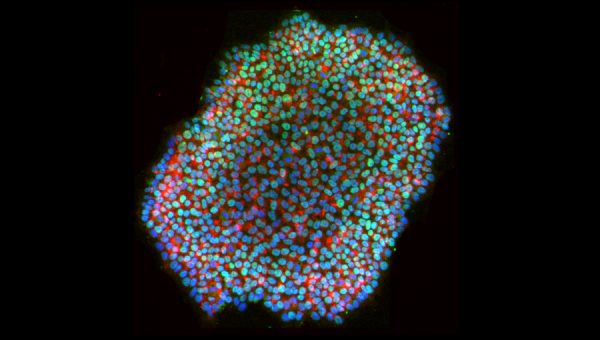
Photo Credit: Image of an Immunofluorescence staining of pluripotency markers Nanog (green) and Tra1-60 (red), with Hoechst 33342(blue) nucleic acid counterstain on MYSM1-KO/F0443.3-C3 IPSC, courtesy of Carol Chen.