MRM Insights: Human Ependymal Cells – The Development of an iPSC protocol

Yue Zhang

Jo Anne Stratton
Every month, in MRM Insights, a member of the MRM Network is writing about stem cells and regenerative medicine from a different perspective. This month, Jo Anne Stratton, Assistant Professor in the Department of Neurology and Neurosurgery at McGill University and Investigator at the Montreal Neurological Institute, and Yue Zhang, Ph.D. candidate in her lab, are discussing how to derive ependymal cells found in the brain from human inducible pluripotent stem cells.
Human Ependymal Cells: The Development of an iPSC protocol
Glial cells are the most abundant and the most diverse cell type in the central nervous system (CNS). Initially believed to only support neurons, glial cells have proven that they play critical roles in brain physiology and pathology. Ependymal cells are one such glia that constitutes the epithelial layer, lining the entire cerebral spinal fluid (CSF)-filled ventricular system, which makes ependymal cells intimately connected with CSF, where they play key roles in regulating CSF dynamics1. This includes regulating CSF flow via motile cilia that beat in a co-ordinated fashion to move CSF throughout the ventricles2,3. Disruption in CSF circulation is detected in neurological diseases, which contributes to an accumulation of neurotoxic waste and aggravates disease processes4–6.
There is a lack of robust modelling systems to explore the role of ependymal cells in health and disease. Indeed, the field relies largely on rodent systems to study these cells, and because mature ependymal cells isolated directly from adult brains fail to survive or proliferate in vitro, the field relies largely on neonatal culturing systems for studying these cells in vitro7. Ependymal cell progenitors (or radial glia) are derived from neonatal rodents and are then induced to differentiate into mature ependymal cells in vitro for their subsequent study7,8. Although useful, rodent ependymal cells do not fully recapitulate human biology, and unfortunately, no robust human assays to study human ependymal cells exist.
Human-induced pluripotent stem cells (iPSCs) are a valuable tool for studying human development, modelling physiological or pathological conditions, and providing a cell source for drug discovery. Protocols to derive a large number of cell types from reprogrammed iPSCs exist, including cells from the CNS like neurons, astrocytes, oligodendrocytes, and microglia9–12, but no such protocols exist for ependymal cells. Ependymal cells are derived from radial glia which are elongated bipolar progenitors that appear in the brain when the telencephalon starts to become a pluristratified epithelium and form a layer between the ventricular zone (VZ) and the preplate13,14. Mimicking the in vivo process of radial glia production, others have generated iPSC protocols to allow for the generation of iPSC radial glia15. Our goal is to extend these protocols to generate ependymal cells, an important step for creating an in vitro human iPSC assay to study ependymal cells in health and disease (Figure 1).
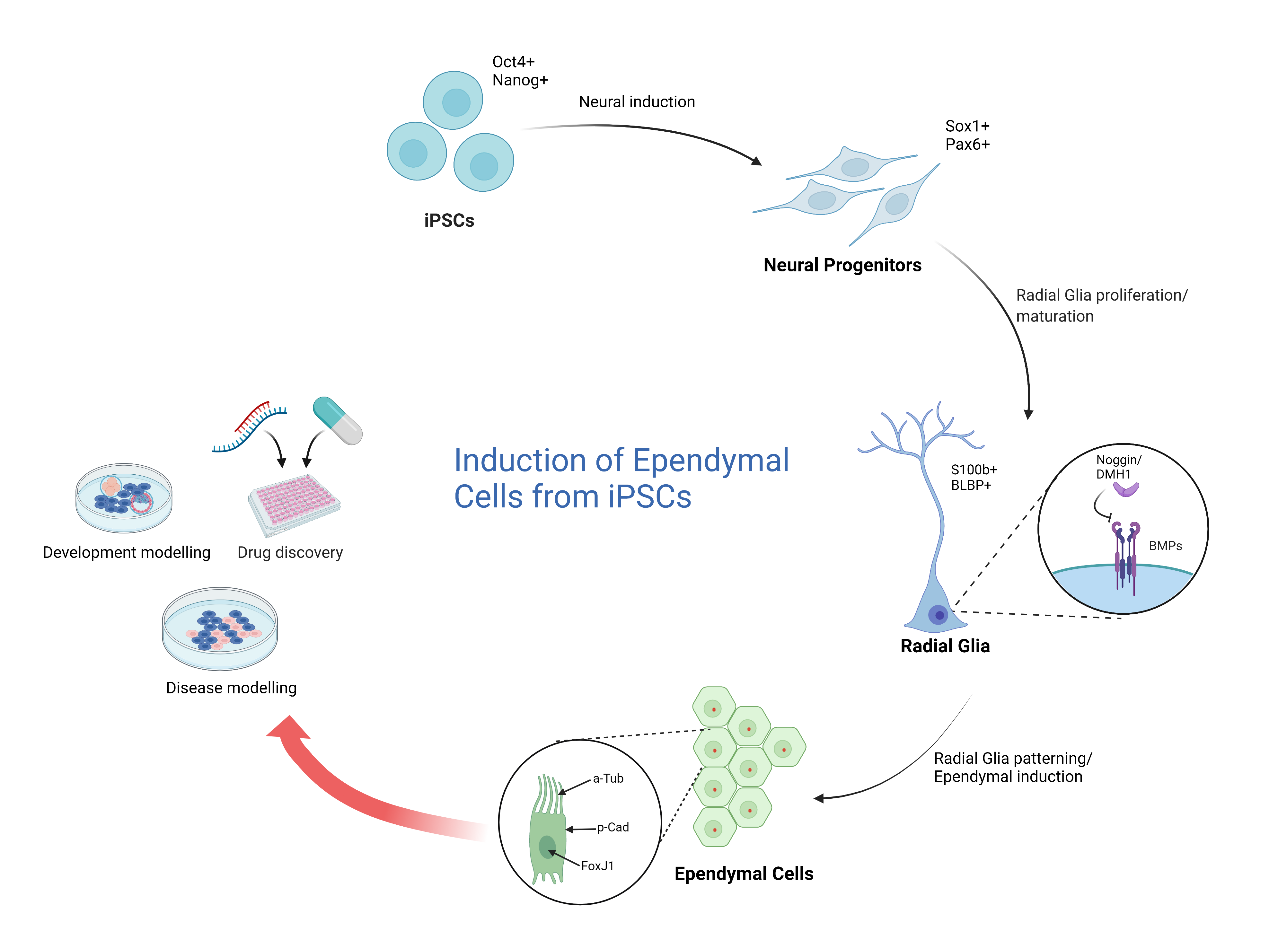
Figure 1
In order to differentiate radial glia into ependymal cells, the defined cues that these cells receive during mid-fetal development must be recapitulated, for example, bone morphogenetic protein (BMP), which is a neuronal growth factor, must be absent. Normally, BMP signalling is prominent during dorsal neural patterning during mid-fetal development but largely absent in the ventral regions where ependymal cells develop16. Indeed, BMP inhibition results in the generation of a large number of centrioles that migrate to the plasma membrane and become the basal bodies of motile cilia17. As such we predict that the inhibition of BMP signalling using BMP antagonists like Noggin or DMH1 in iPSC radial glia will result in the formation of cilia important for ependymal cell maturation18. In addition, we also aim to withdraw all mitogens and add a neddylation inhibitor, MLN-4924, that stabilizes the transcription factor, Foxj1 (forkhead box protein J 1) – the master transcription factor involved in ciliogenesis19. MLN-4924 stabilizes FOXJ1 protein by inhibiting its degradation through the proteasomal pathway, resulting in the generation of stable ependymal cell populations in vitro3. With these modifications, we hope to create a robust iPSC ependymal cell culturing system that we will characterize using immunofluorescent staining, transcriptional phenotyping, and functional assays.
Developing a method for generating ependymal cell cultures from human-derived iPSCs is unprecedented. It will help us better understand the development of ependymal cells as well as support our research in assessing ependymal cells in health and disease20.
References
1. MacDonald, A. et al. Single Cell Transcriptomics of Ependymal Cells Across Age, Region and Species Reveals Cilia-Related and Metal Ion Regulatory Roles as Major Conserved Ependymal Cell Functions. Front. Cell. Neurosci. 15, (2021).
2. Del Bigio, M. R. The ependyma: a protective barrier between brain and cerebrospinal fluid. Glia 14, 1–13 (1995).
3. Abdi, K. et al. Uncovering inherent cellular plasticity of multiciliated ependyma leading to ventricular wall transformation and hydrocephalus. Nat. Commun. 9, (2018).
4. Reeves, B. C. et al. Glymphatic System Impairment in Alzheimer’s Disease and Idiopathic Normal Pressure Hydrocephalus. Trends Mol. Med. 26, 285–295 (2020).
5. Louveau, A. et al. Understanding the functions and relationships of the glymphatic system and meningeal lymphatics. J. Clin. Invest. 127, 3210–3219 (2017).
6. Ma, Q., Ineichen, B. V., Detmar, M. & Proulx, S. T. Outflow of cerebrospinal fluid is predominantly through lymphatic vessels and is reduced in aged mice. Nat. Commun. 8, (2017).
7. Shah, P. T. et al. Single-Cell Transcriptomics and Fate Mapping of Ependymal Cells Reveals an Absence of Neural Stem Cell Function. Cell 173, 1045-1057.e9 (2018).
8. Gabrion, J. B. et al. Ependymal and choroidal cells in culture: Characterization and functional differentiation. Microsc. Res. Tech. 41, 124–157 (1998).
9. Hasselmann, J. & Blurton-Jones, M. Human iPSC-derived microglia: A growing toolset to study the brain’s innate immune cells. Glia 68, 721–739 (2020).
10. Emdad, L., D’Souza, S. L., Kothari, H. P., Qadeer, Z. A. & Germano, I. M. Efficient differentiation of human embryonic and induced pluripotent stem cells into functional astrocytes. Stem Cells Dev. 21, 404–410 (2012).
11. TCW, J. et al. An Efficient Platform for Astrocyte Differentiation from Human Induced Pluripotent Stem Cells. Stem cell reports 9, 600–614 (2017).
12. Hu, B. Y. & Zhang, S. C. Differentiation of spinal motor neurons from pluripotent human stem cells. Nat. Protoc. 4, 1295–1304 (2009).
13. Malatesta, P., Appolloni, I. & Calzolari, F. Radial glia and neural stem cells. Cell Tissue Res. 331, 165–178 (2008).
14. Redmond, S. A. et al. Development of Ependymal and Postnatal Neural Stem Cells and Their Origin from a Common Embryonic Progenitor. Cell Rep. 27, 429-441.e3 (2019).
15. Duan, L., Peng, C.-Y., Pan, L. & Kessler, J. A. Human Pluripotent Stem Cell-Derived Radial Glia Recapitulate Developmental Events and Provide Real-Time Access to Cortical Neurons and Astrocytes. Stem Cells Transl. Med. 4, 437–447 (2015).
16. Wurst, W., Bally-Cuif, L. & Bally-Cuif, L. Neural plate patterning: Upstream and downstream of the isthmic organizer. Nat. Rev. Neurosci. 2001 22 2, 99–108 (2001).
17. Delgehyr, N. et al. Ependymal cell differentiation, from monociliated to multiciliated cells. Methods Cell Biol. 127, 19–35 (2015).
18. Hiraoka, K., Inada, H., Yanai, K. & Osumi, N. Bone Morphogenetic Proteins Inhibit Ciliogenesis of Ependymal Cells in Vitro. Tohoku J. Exp. Med. 252, 199–208 (2020).
19. Jacquet, B. V. et al. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development 136, 4021–4031 (2009).
20. Hatrock, D., Caporicci-Dinucci, N. & Stratton, J. Ependymal cells and multiple sclerosis: proposing a relationship. Neural Regen. Res. 15, 263–264 (2020).
Photo Credit: courtesy of Dale Hatrock.
